Correlation Calculator for Metabolomics Data
The Correlation Calculator is a standalone Java application providing various methods of calculating pairwise correlations among repeatedly measured entities. It is designed for use with quantitative metabolite measurements such as MS data on a set of samples. The workflow allows inspection and/or saving of results at various stages, and the final correlation results can be dynamically imported into MetScape (version 3.1 or higher) as a correlation network.
Download the Correlation Calculator
Download sample input file: aminoacids.csv
Input File Format
For this release, input files are required to be in CSV format. Input files should be tables of measurements of metabolites across multiple samples. Metabolites must be labelled, but sample labels are optional. Samples may be in either rows or columns. The example input file does not have labelled samples, and its samples are in rows.
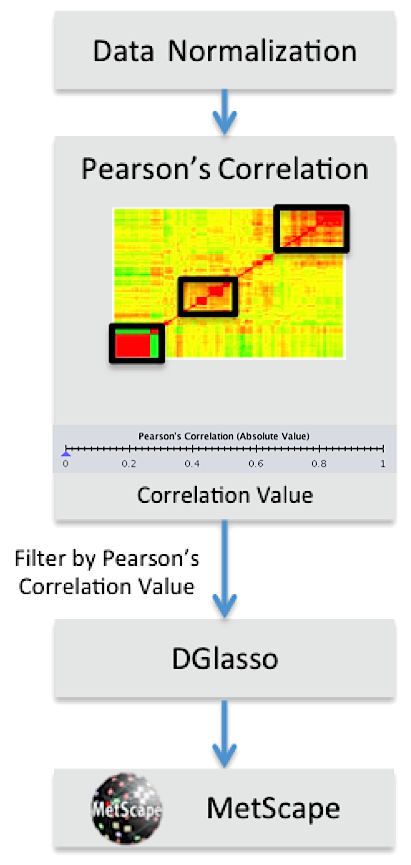
Correlation Calculator Workflow
After a data file has been imported, users have the option of normalizing
the data using a log transformation and/or autoscaling.
Pearson's correlation coefficients are useful both to get an overview of
correlation structures and as a way of filtering large data sets.
Results may be viewed as a static heatmap in PDF format or exported to
a file or an interactive heatmap viewer such as TreeView.
A slider is provided for filtering data sets based on a Pearson's correlation
threshold. Metabolites with no Pearson's correlations above the threshold
value are excluded from subsequent analyses.
Partial correlations can be calculated on the filtered data using a De-sparsified
Graphical lasso (DGlasso) method developed by our collaborators George
Michailidis and Sumanta Basu. The DGlasso method is particularly useful when
the number of metabolites exceeds the number of samples in the data set.
Partial correlation results can then be viewed interactively in
MetScape as a correlation network.